前言
地理隔离是物种遗传多样性的基础。不同地理分布的种群往往具有不同的基因分布。我们使用水稻某基因的单倍型和中国各省份降水量数据对该基因的单倍型进行可视化。
单倍型生成
利用genehapR包将vcf转换为单倍型数据
suppressWarnings(suppressMessages(library(tidyverse)))
suppressWarnings(suppressMessages(library(geneHapR)))
vcf <- import_vcf("C:/Users/wpf/Desktop/project/Popgenetic/whj/data/zfpvg.vcf")## Scanning file to determine attributes.
## File attributes:
## meta lines: 17
## header_line: 18
## variant count: 2
## column count: 4735
##
Meta line 17 read in.
## All meta lines processed.
## gt matrix initialized.
## Character matrix gt created.
## Character matrix gt rows: 2
## Character matrix gt cols: 4735
## skip: 0
## nrows: 2
## row_num: 0
##
Processed variant: 2
## All variants processedhap <- vcf2hap(vcf) %>%
filter(str_starts(Hap, "H")) %>%
mutate(Hap = if_else(Hap == "H001", "A", "B"),
taxa = str_replace(Accession, "\\.", "-")) %>%
select(taxa, Hap)## Extracting gt element GT# 选择中国的省份,并将部分省份合并
chinapro <- c("Jiangsu", "Hunan", "Taiwan", "Jiangxi", "Guangdong", "Sichuan", "Anhui", "Zhejiang", "Yunnan",
"Hubei", "Fujian", "Guizhou", "Liaoning; China", "yunnan", "Heilongjiang", "Hebei", "Guangxi", "Henan",
"Shanghai", "Xizang", "Shanxi", "Liaoning", "Guangxi; China", "Hainan", "Tianjin", "Hong Kong", "Beijing",
"Jilin")
key <- tibble(
name = c("Jiangsu", "Hunan", "Taiwan", "Jiangxi", "Guangdong", "Sichuan", "Anhui", "Zhejiang", "Yunnan",
"Hubei", "Fujian", "Guizhou", "Heilongjiang", "Hebei", "Guangxi", "Henan","Shanghai", "Xizang",
"Shanxi", "Liaoning", "Hainan", "Tianjin", "Beijing", "Jilin"),
value = c("Jiangsu", "Hunan", "Taiwan", "Jiangxi", "Guangdong", "Sichuan", "Anhui", "Zhejiang", "Yunnan",
"Hubei", "Fujian", "Guizhou", "Jilin", "Hebei", "Guangxi", NA,"Jiangsu", NA,
NA, "Jilin", NA, "Hebei", "Hebei", "Jilin")
) %>%
pull(value, name = name)
group <- readxl::read_excel("C:/Users/wpf/Desktop/project/Popgenetic/VarMap/taxainfo.xlsx") %>%
select(`Cultivar ID`, Location) %>%
rename(taxa = `Cultivar ID`) %>%
filter(Location %in% chinapro) %>%
mutate(Location = case_when(Location == "Liaoning; China" ~ "Liaoning",
Location == "yunnan" ~ "Yunnan",
Location == "Guangxi; China" ~ "Guangxi",
TRUE ~ Location)) %>%
mutate(Location = recode(Location, !!!key, .default = NA_character_)) %>%
na.omit() %>%
left_join(hap, by = "taxa") %>%
count(Location, Hap) %>%
pivot_wider(names_from = Hap,
values_from = n,
values_fill = 0)
location <- group %>%
pull(Location)
info <- readxl::read_excel("C:/Users/wpf/Desktop/project/Popgenetic/VarMap/Raw Data submission.xls", skip = 11, col_names = FALSE) %>%
select(3, 4, 5) %>%
`colnames<-`(c("Location", "lat", "long")) %>%
filter(Location %in% location) %>%
distinct(Location, .keep_all = TRUE) ## New names:
## • `` -> `...1`
## • `` -> `...2`
## • `` -> `...3`
## • `` -> `...4`
## • `` -> `...5`
## • `` -> `...6`
## • `` -> `...7`
## • `` -> `...8`
## • `` -> `...9`group2 <- group %>%
left_join(info, by = "Location") %>%
distinct(Location, .keep_all = TRUE) %>%
select(long, lat, Location, A, B) %>%
mutate(radius = (A + B)/10,
long = as.numeric(long),
lat = as.numeric(lat))
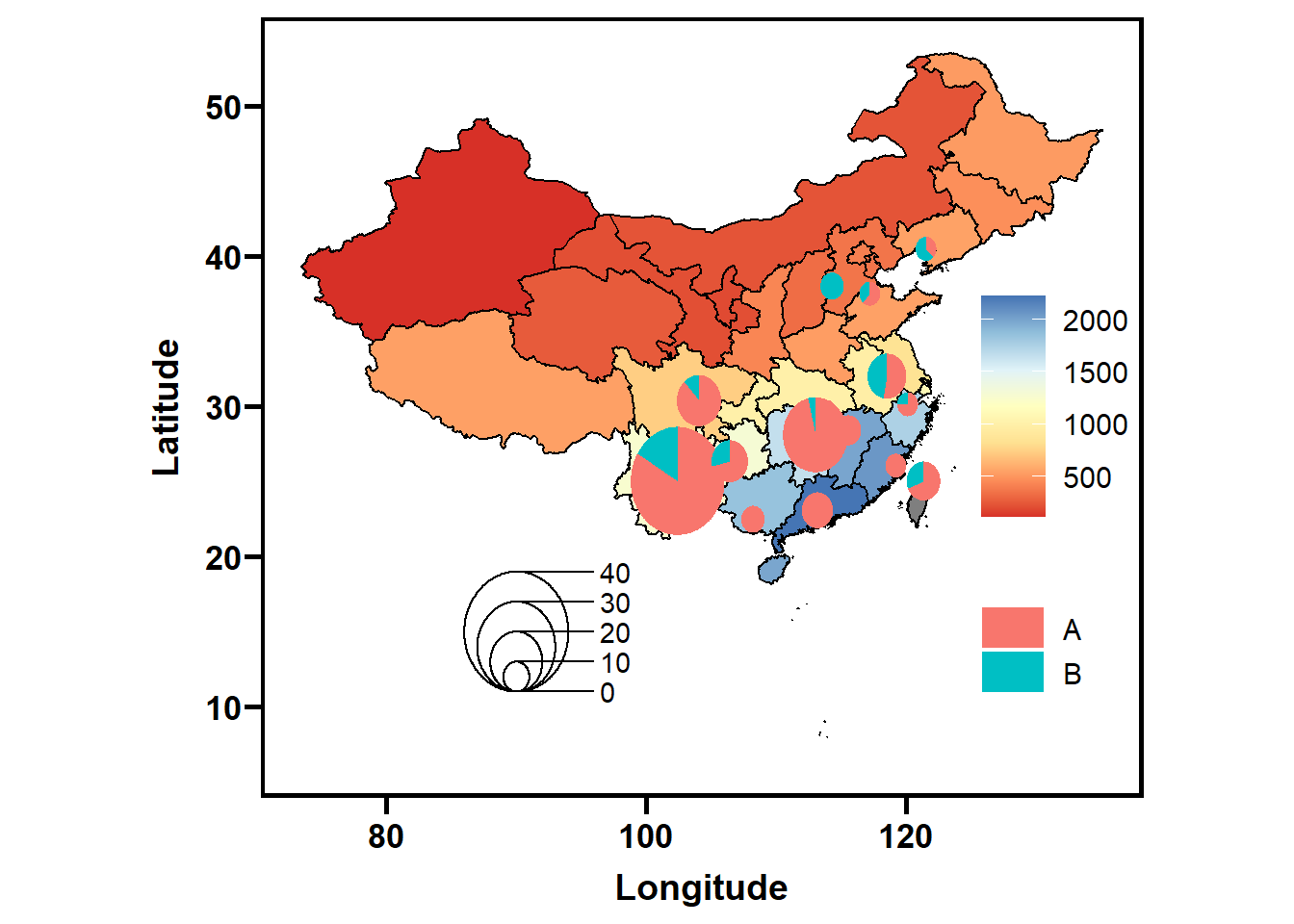
# 单倍型输入格式如下
group2## # A tibble: 15 × 6
## long lat Location A B radius
## <dbl> <dbl> <chr> <int> <int> <dbl>
## 1 117. 37.5 Anhui 5 3 0.8
## 2 119. 26.0 Fujian 8 0 0.8
## 3 113. 23.1 Guangdong 12 0 1.2
## 4 108. 22.5 Guangxi 9 0 0.9
## 5 106. 26.4 Guizhou 10 4 1.4
## 6 114. 38.0 Hebei 0 9 0.9
## 7 114 30 Hubei 4 0 0.4
## 8 113 28.1 Hunan 24 1 2.5
## 9 118. 32.0 Jiangsu 8 7 1.5
## 10 116. 28.4 Jiangxi 10 0 1
## 11 122. 40.5 Jilin 3 5 0.8
## 12 104. 30.4 Sichuan 15 2 1.7
## 13 121. 25.0 Taiwan 9 4 1.3
## 14 102. 25.0 Yunnan 30 6 3.6
## 15 120. 30.1 Zhejiang 6 2 0.8可视化
利用chinamap和scatterpie包共同完成可视化,其中ggnewscale包提供了二次着色的可能。
suppressWarnings(suppressMessages(library(chinamap)))
suppressWarnings(suppressMessages(library(scatterpie)))
suppressWarnings(suppressMessages(library(ggnewscale)))
suppressWarnings(suppressMessages(library(ggprism)))
cn <- get_map_china() ## Loading required package: sp## Warning: package 'sp' was built under R version 4.3.1## The legacy packages maptools, rgdal, and rgeos, underpinning the sp package,
## which was just loaded, will retire in October 2023.
## Please refer to R-spatial evolution reports for details, especially
## https://r-spatial.org/r/2023/05/15/evolution4.html.
## It may be desirable to make the sf package available;
## package maintainers should consider adding sf to Suggests:.
## The sp package is now running under evolution status 2
## (status 2 uses the sf package in place of rgdal)##
## Attaching package: 'sp'## The following object is masked from 'package:scatterpie':
##
## recenter## Regions defined for each Polygonsrainfall <- readxl::read_excel("C:/Users/wpf/Desktop/project/Visualization/whj/rainfall2.xlsx")
tmp <- cn %>%
left_join(rainfall, by = "province") %>%
as_tibble()
ggplot() +
geom_map(aes(long, lat, map_id = id, fill = Rain), color = "black", map = tmp, data = tmp) +
scale_fill_distiller(palette = "RdYlBu", direction = 1) +
coord_quickmap() +
new_scale_fill() +
geom_scatterpie(aes(x = long, y = lat, group = Location, r = radius),
data = group2,
cols = LETTERS[1:2],
color = NA) +
theme_prism(border = TRUE, base_rect_size = 1.5) +
theme(legend.position = c(0.9, 0.4))+
geom_scatterpie_legend(group2$radius, x= 90, y = 15, labeller = function(x) {10*x}) +
labs(x = "Longitude", y = "Latitude")## Warning: Unknown or uninitialised column: `region`.## Warning in geom_map(aes(long, lat, map_id = id, fill = Rain), color = "black",
## : Ignoring unknown aesthetics: x and y